I was recently notified of this: our article ‘DNA barcodes for 1/1000 of the Animal Kingdom‘ is one of the top ten most cited Biology Letters articles for 2010. Here’s one good paper that cited it, a recent PLoS ONE paper by Axel Hausmann and colleagues.
Archive for the ‘Publications’ Category
One more PLoS ONE paper online
Another chapter from my thesis came out in PLoS ONE recently. It’s a brief study that compiles and assesses a nearly complete barcode library for the geometrids of BC (for a great overview of this family in BC, see p75 of Scudder and Cannings 2007). If you’re curious, the only species on our checklist that we didn’t successfully barcode was Hydrelia brunneifasciata (Packard)). Grab the open-access article here or from my ‘publications’ page, and the reference is as follows:
deWaard, J.R., Hebert, P.D.N., and Humble, L.M. (2011). A comprehensive DNA barcode library for the looper moths (Lepidoptera: Geometridae) of British Columbia, Canada. PLoS ONE 6: e18290.
PLoS ONE paper online
A paper that 10 colleagues and I wrote is now online at PLoS ONE. It develops and tests a partial global barcode library for surveillance of tussock moths of the genus Lymantria. You can find it here and the citation follows:
deWaard, J.R., Mitchell, A., Keena, M.A., Gopurenko, D., Boykin, L.M., Armstrong, K.F., Pogue, M.G., Lima, J., Floyd, R., Hanner, R.H. and Humble, L.M. 2010. Towards a global barcode library for Lymantria (Lepidoptera: Lymantriinae) tussock moths of biosecurity concern. PLoS ONE 5: e14280. doi:10.1371/journal.pone.0014280
Update: this is now part of a collection on the PLOS One site titled “Proceedings of the Third International Barcode of Life Conference, Mexico City“.
New article in press
Our manuscript describing the detection of the juniper pug moth (Eupithecia pusillata) in North America has been accepted for publication in the Journal of the Entomological Society of British Columbia. The abstract is here and the citation is:
deWaard, J.R., Humble, L.M., and Schmidt, B.C.S. 2010. DNA barcoding identifies the first North American records of the Eurasian moth, Eupithecia pusillata (Lepidoptera: Geometridae). Journal of the Entomological Society of BC. In press.
Another milestone
Not long after the milestone of 1 million barcodes was reached in time for the official launch of the International Barcode of Life Project, our original barcoding paper has now reached 1000 citations according to ISI Web of Knowledge.
Stumbled upon this…
Our original barcoding paper is now the 6th most cited paper ever for Proceedings B. Surprisingly, it’s cited more than Gould and Lewontin’s classic 1979 paper on the ‘Spandrels of San Marco’. Our 2nd barcoding paper is 47th, and they’re the two most recent in the top 50 list.
New article in Biological Invasions online
Floyd, R., Lima, J., deWaard, J.R., Humble, L.R. and Hanner, R.H. (2010). Common goals: incorporating DNA barcoding into international protocols for identification of arthropod pests. Biological Invasions. In press. [full text pdf] [website].
Abstract: The globalization of commerce carries with it significant biological risks concerning the spread of harmful organisms. International Standards for Phytosanitary Measures (ISPM) No. 27, ‘‘Diagnostic Protocols for Regulated Pests’’, sets out the standards governing protocols for the detection and identification of plant pest species. We argue that DNA barcoding—the use of short, standardized DNA sequences for species identification—is a methodology which should be incorporated into standard diagnostic protocols, as it holds great promise for the rapid identification of species of economic importance, notably arthropods. With a well-defined set of techniques and rigorous standards of data quality and transparency, DNA barcoding already meets or exceeds the minimum standards required for diagnostic protocols under ISPM No. 27. We illustrate the relevance of DNA barcoding to phytosanitary concerns and advocate the development of policy at the national and international levels to expand the scope of barcode coverage for arthropods globally.
DNA barcodes for 1/1000 of the Animal Kingdom. Biology Letters
Hebert, P.D.N., deWaard, J.R., and Landry, J.-F. DNA barcodes for 1/1000 of the Animal Kingdom. Biology Letters 6: 359-362 [abstract pdf] [full text pdf] [website]
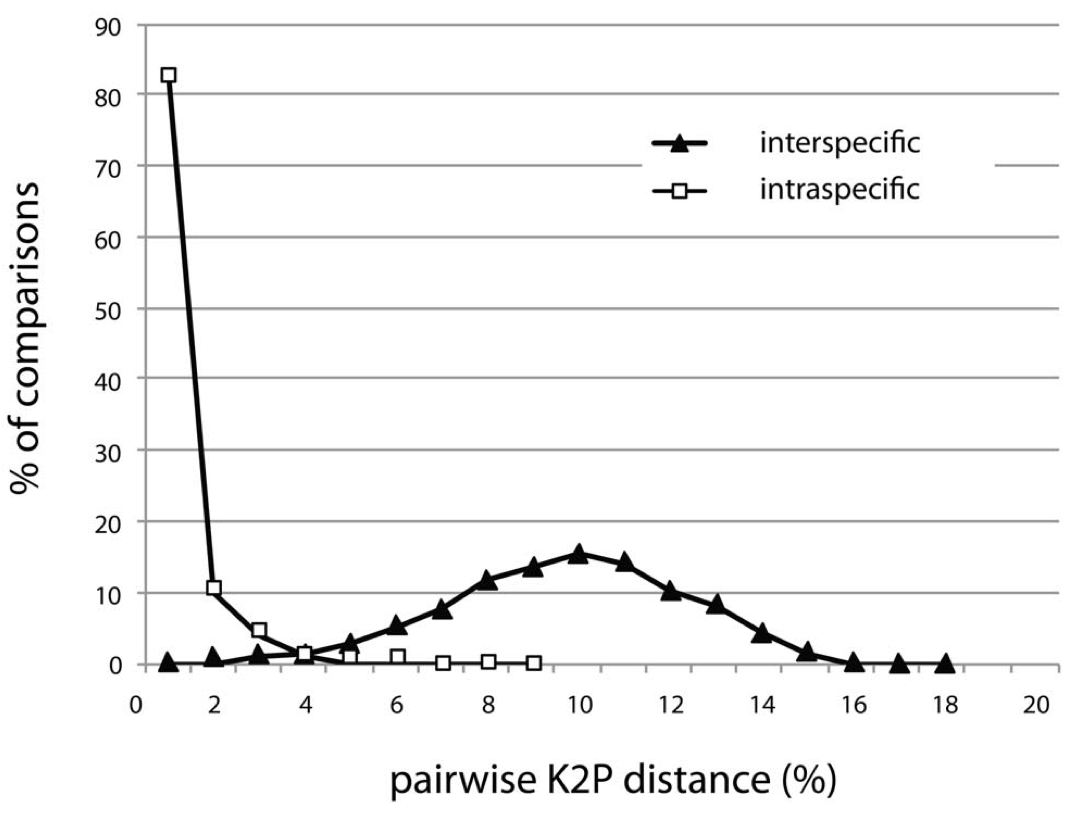
Abstract: This study reports DNA barcodes for more than 1300 Lepidoptera species from the eastern half of North America, establishing that 99.5% of these species possess diagnostic barcode sequences. Intraspecific divergences averaged just 0.43% among this assemblage, but most values were lower. The mean was elevated by deep barcode divergences (>2%) in nearly 5% of the species, often involving the sympatric occurrence of two barcode clusters. A few of these cases have been analyzed in detail, revealing species overlooked by the current taxonomic system. This study also provided a large-scale test of the extent of regional divergence in barcode sequences, indicating that geographic differentiation is small, even when comparisons involve populations as much as 2800 km apart. The present results affirm that a highly effective system for the identification of Lepidoptera can be built with few records per species because of the limited intra-specific variation. As most terrestrial and marine taxa possess patterns of population structure similar to those in Lepidoptera, an effective DNA-based identification system can be developed with modest effort.
Testing the utility of mitochondrial DNA (COI) sequences for the identification and phylogeny of New Zealand caddisflies (Trichoptera). New Zealand Journal of Marine and Freshwater Research
Hogg, I.D., Smith, B.J., Banks, J.C., deWaard, J.R., and Hebert, P.D.N. (2009). Testing the utility of mitochondrial DNA (COI) sequences for the identification and phylogeny of New Zealand caddisflies (Trichoptera). New Zealand Journal of Marine and Freshwater Research 43: 1137-1146. [abstract pdf] [full text pdf] [website]
Abstract: We tested the hypothesis that cytochrome c oxidase subunit 1 (COI) sequences would successfully discriminate recognised species of New Zealand caddisflies. We further examined whether phylogenetic analyses, based on the COI locus, could recover currently recognised superfamilies and suborders. COI sequences were obtained from 105 individuals representing 61 species and all 16 families of Trichoptera known from New Zealand. No sequence sharing was seen between members of different species and congeneric species showed from 2.3 – 19.5% divergence. Sequence divergence among members of a species was typically low (mean = 0.7%; range 0-8.5%), but two species showed intraspecific divergences in excess of 2%. Phylogenetic reconstructions based on COI were largely congruent with prior conclusions based on morphology, although the sequence data did not support placement of the purse-cased caddisflies (Hydroptilidae) within the uncased caddisflies, and in particular, the Rhyacophiloidea. We conclude that sequence variation in the COI gene locus is not only a useful tool for the identification of New Zealand caddisfly species, but that it can provide preliminary phylogenetic inferences. Further work is needed to ascertain the significance of the few cases of high intra-specific divergence and to determine if any cases of sequence sharing will be detected with larger sample sizes.
In the dark in a large urban park: DNA barcodes illuminate cryptic and introduced moth species. Biodiversity and Conservation
deWaard, J.R., Landry, J.-F., Schmidt, B.C., Derhousoff, J., McLean, J.A. and Humble, L.M. (2009). In the dark in a large urban park: DNA barcodes illuminate cryptic and introduced moth species. Biodiversity and Conservation 18:3825–3839. [full text pdf] [website]
Abstract: To facilitate future assessments of diversity following disturbance events, we conducted a first level inventory of nocturnal Lepidoptera in Stanley Park, Vancouver, Canada. To aid the considerable task, we employed high-throughput DNA barcoding for the rough sorting of all material and for tentative species identifications, where possible. We report the preliminary species list of 190, the detection of four new exotic species (Argyresthia pruniella, Dichelia histrionana, Paraswammerdamia lutarea, and Prays fraxinella), and the potential discovery of two cryptic species. We describe the magnitude of assistance that barcoding presents for faunal inventories, from reducing specialist time to facilitating the detection of native and exotic species at low density.